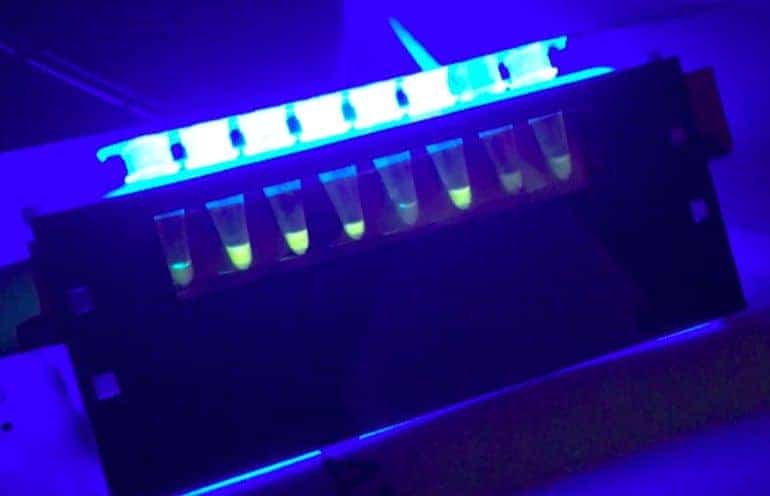
Neither the diagnosis of cases of SARS-CoV-2 nor the detection of environmental contamination by the virus is yet straightforward. Complicated molecular amplification techniques, which require extensive infrastructure and biosafety procedures, prevent people from knowing whether the virus is actually present in their environment.
The “Do-It-Together SARS CoV-2 Detective” project (more information here) will develop a strategy similar to that used for the “GMO Detective“This is the only way to detect the virus responsible for Covid-19. The #CoronaDetective is expected to be highly specific, not only without complicated equipment, but also with a simple +/- reading. In addition, controls to ensure sensitive detection, with no false positives or negatives, are intrinsic to this solution. The final product, tube strips with dry reagents, could be easily supplied anywhere, without dependence on the cold chain; and monitoring tests could be carried out by ordinary people, with no training in medicine or biology, just a certain ability to follow simple instructions and common sense. In the future, the project will try out “multiplexed” fluorescent detection, using an inexpensive (even DIY) detector, but with a partner project (Slack #proj-neb-lamp-test), it would also be nice to find out whether a simpler color change reaction will be sufficient to obtain valid results.
Human or clinical samples should only be analyzed in environments with access to appropriate biosafety facilities, of course. This project is fully compliant with the OpenCovid-19 initiative's biosafety and biosecurity guidelines, and is fortunate to have appropriate laboratories for all levels of validation. Thanks to freely available information, targeted regions of the virus have already been analyzed, and one has even proved identifiable by team members using these molecular techniques. Our aim now is to optimize the components and develop a validated method that can be used by everyone.
This project is part of the’OpenCovid19 JOGL initiative, which aims to collectively develop free, inexpensive, safe and easy-to-use tools and methodologies to combat the COVID-19 pandemic, and is being developed as part of the Covid19 Diagnostic and Detection Challenge. It was recently funded by the first wave of beneficiaries of the JOGL open peer-review micro-grant scheme. Everyone can play a part!
Together, open participatory research can help solve today's problems! We welcome your help!
How to contribute
You can reach us via this Slack channel: #proj-nucleic-acid-amplification of l’initiative JOGL Open Covid-19. All this is discussed in : #proj-nucleic-acid-amplification, and more in #chlg-detection-diagnosis. Related work on different RNA extraction methods can be found in: #proj-nucleic-acid-amplification : #proj-rna-extraction.
Further work is underway to produce and test open source versions of the enzymes needed in #proj-enzyme-production #freegenes, which are linked to : https://openbioeconomy.org/projects/open-enzyme-collections/ and https://biobricks.org/freegenes/
Ali Bektas, a collaborator at the Oakland Genomics Center in California, is actively trying to put it all together in a small cartridge, see https://app.jogl.io/project/187. Other people are also working on similar methods https://app.jogl.io/project/163. These two projects were also funded by the first wave of beneficiaries of the JOGL open peer-review micro-grant scheme.